🚀 I am thrilled to share our advancements in therapeutic target discovery with Mantis-ML 2.0, now published in Science Advances! 🎉
#AstraZeneca #AI #Genomics
Mantis-ML 2.0 is an ensemble #ML framework designed to comprehensively harness the wealth of existing scientific knowledge and AstraZeneca’s Biological Insights Knowledge Graph to identify promising therapeutic targets for diseases.
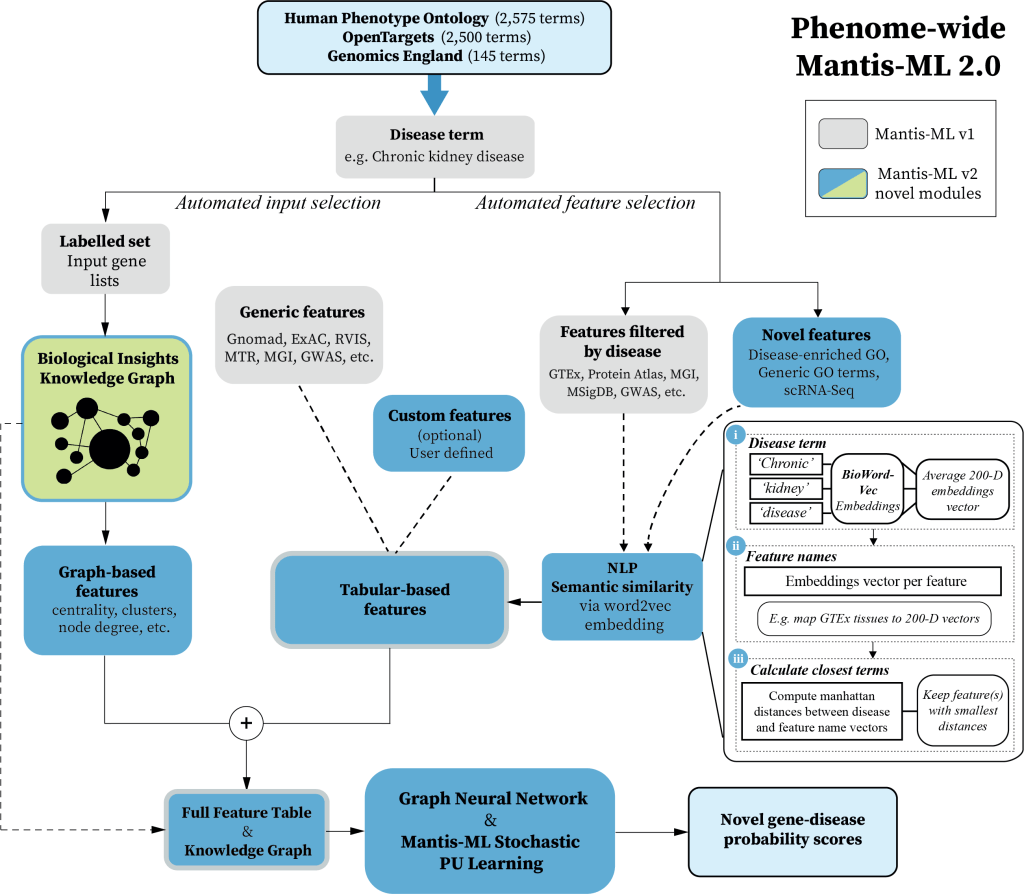
📈💪 Improved Performance
Mantis-ML 2.0 represents a significant leap forward from previous iterations, by integrating #KnowledgeGraphs, #GraphNeuralNetworks and automated selection of disease relevant features for training. The upgraded models compared to our 2020 Mantis-ML publication (Vitsios & Petrovski, AJHG) boast a significant enhancement in classification power — a 6.9% boost on average! With a median ROC AUC score of 0.90 over 5,220 diseases.
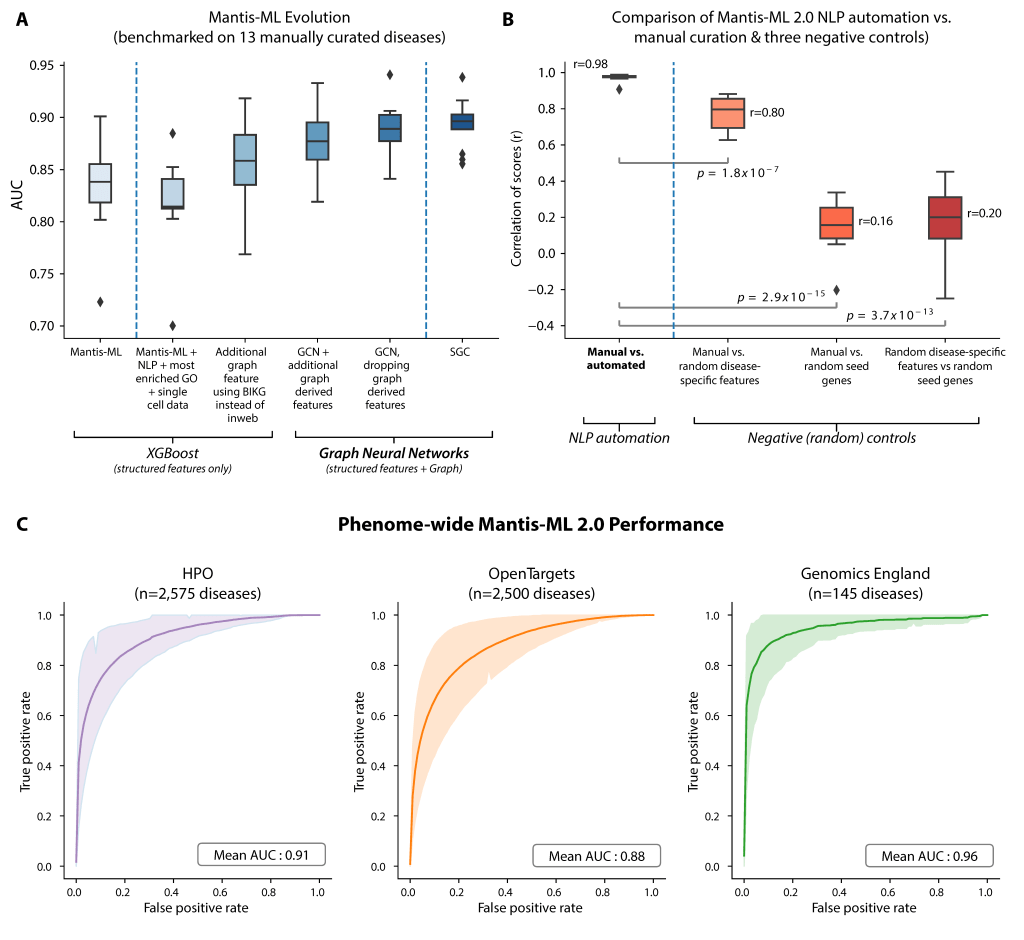
💡 Why Mantis-ML 2.0?
Mantis-ML 2.0 uses advanced graph neural networks to grasp the networked structure of multi-dimensional gene-level annotation data. It’s trained on numerous balanced datasets via a robust semi-supervised learning framework, which enables it to provide detailed gene-disease probabilities across the whole human exome.
Importantly, we cross-overlap Mantis-ML 2.0 predictions with associations from an independent #UK Biobank phenome-wide association study (#PheWAS), providing a stronger form of triaging and mitigating against underpowered PheWAS associations.
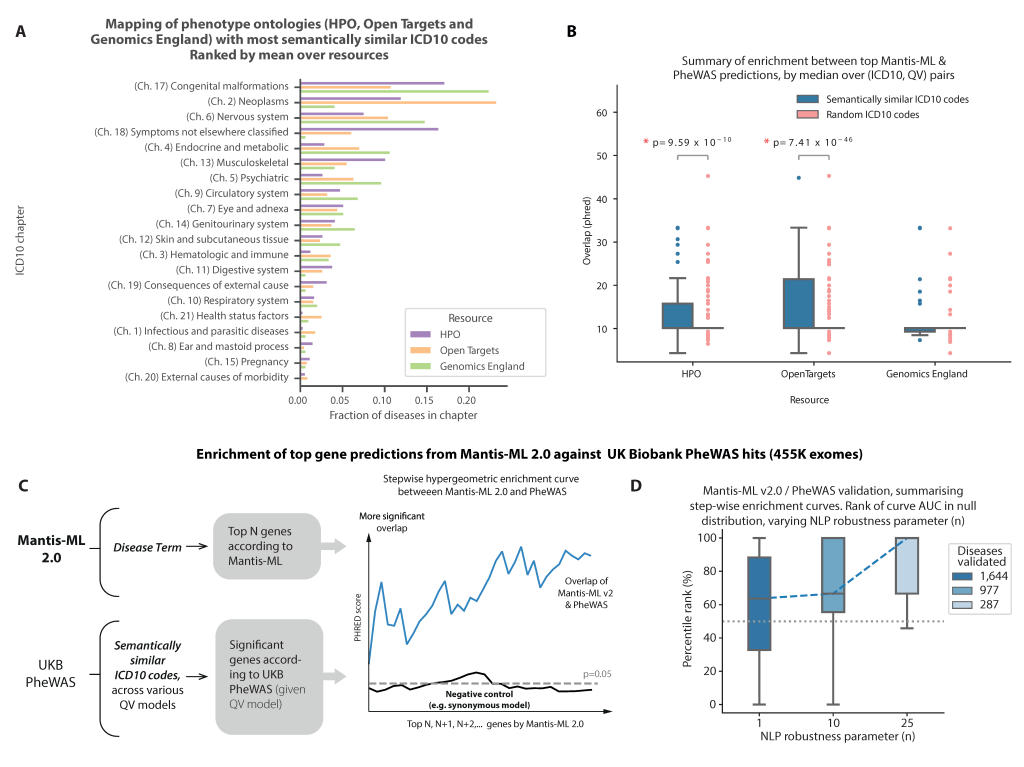
❓💡How to access Mantis-ML 2.0 results?
We have trained Mantis-ML 2.0 across > 5,000 diseases and expose our results in a public interactive portal: http://mantisml.public.cgr.astrazeneca.com
Researchers may also train their own versions of Mantis-ML 2.0 disease-specific models, leveraging internal knowledge graphs and/or additional feature sets, using our open source tool:
We hope our published method and results will offer novel insights in therapeutic target discovery and enable a series of other innovations in the domain. 🌟

Leave a comment